The role of tobacco smoke-induced changes of the microbiome in shaping risk for IBD

Introduction
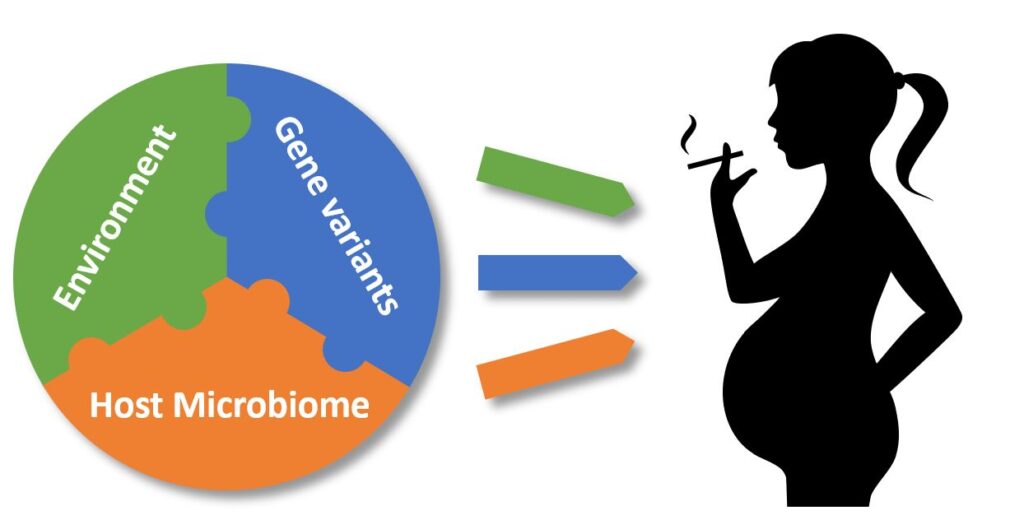
The incidence of IBD has been dramatically increasing over the past decades, which cannot be explained by genetics alone. Current pathophysiological concepts suggest a triangular interaction of IBD risk genes such as IL10, the hosts´ microbiome and environmental factors where tobacco smoking is an established risk factor for developing IBD. Of concern, maternal smoking during pregnancy is also associated with an increased disease risk for IBD in offspring indicating that epigenetic and/or microbial perturbations contribute to development of IBD in later life already in utero.
Aims
We therefore aim to understand how smoking affects the gut microbiome and epigenetics in the context of IBD risk genes during both early life and in adulthood in order to create the prerequisites for novel preventative and/or therapeutic strategies for IBD targeting the microbiome.
We here aim
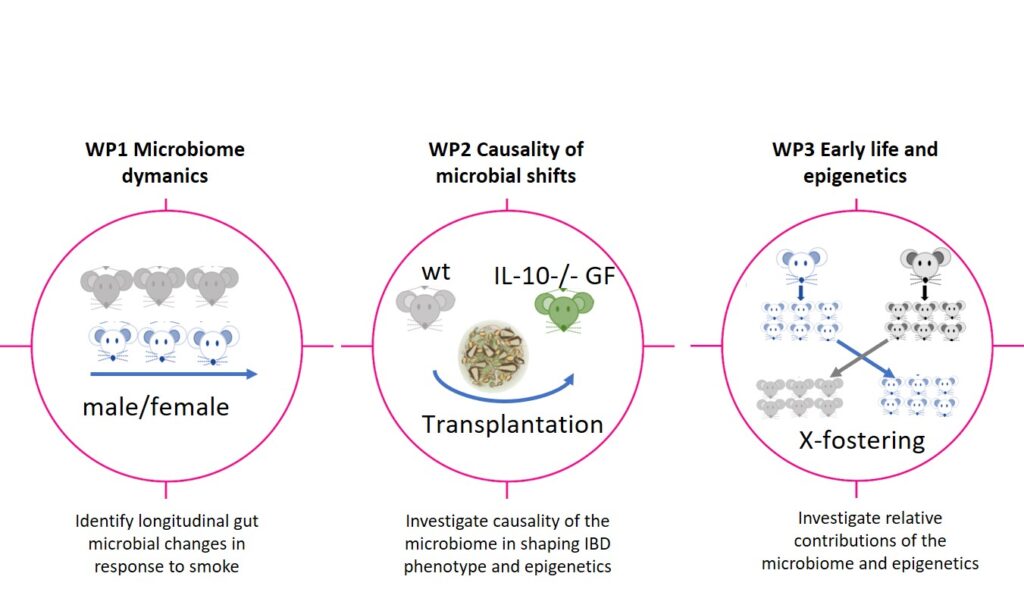
- To monitor dynamics and modulations of the microbiome in response to cigarette smoke exposure. Addressing this objective will create a dynamic picture of how smoking modulates the microbiome and its functions and define the individual stages of an altered microbiome
- to assess the potential causality by transplanting an at-risk microbiome into germ-free mice. This key objective will help the applicants to assess the potential causality of a smoke-shifted microbiome for IBD development. The transplantation experiment planned here is a powerful tool to discriminate between observations which are consequences of smoking and microbiome modulations which are potentially causative for the disease risk, and
- to determine the relative contribution of changes in epigenetic profiles versus the microbiome during maternal smoking to disease susceptibility. Addressing this objective will finally help us to better understand the impact of parental smoking as a driving force of modulating the microbiome. Finally, employing epigenetic and transcriptome analysis, this objective will deliver a picture of the molecular consequences in the host, resulting from parental smoking. This may have considerable consequences for public health policies a as parental behaviour could influence the disease risk of offspring many years later.
Outlook
The project will generate novel knowledge on potential smoking induced microbial shifts in early life and adulthood. As an outlook, proof-of-principle intervention studies will be performed in humanized mouse models using faecal material from at-risk individuals, which will help to guide human intervention strategies in collaboration with P1 and P9. In collaboration with the paediatrics department of the University Clinic of Schleswig-Holstein (UKSH), Kiel, we plan to assess the impact of parental smoking on the microbiome of children.
Researchers
Other important members of P4
- Natalie El-Mehrie, PhD
- Janin Braun, techncian
- Eistine Boateng, bioinformatician PhD
- Dr. Sören Franzenburg, leader of the NGS laboratory (Link CCGA)
- Tanja Klostermeier, technician
- Dr. Neha Mishra, bioinformatician




