Metabolomics, microbial community modelling and data integration
Introduction
This Z-project will provide a collaborative platform within miTarget to investigate the role of metabolism in microbiome-host-interactions in the context of inflammatory bowel diseases (IBD). The Z-project comprises metabolomics (Heinzmann group, Munich) and metabolic modelling (Kaleta group, Kiel). Both approaches facilitate a complementary view on the role of the microbiome in IBD. Metabolomics will identify disease-associated shifts in metabolites of the microbiome and the host. Metabolic modelling will mechanistically elucidate the contribution of microbiome-host-interactions to disease.
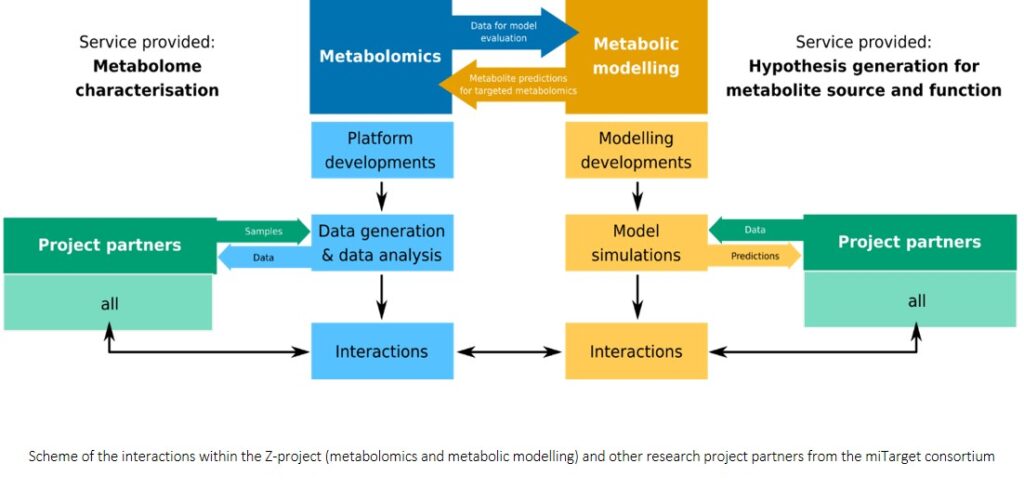
Non-targeted metabolomics techniques available at Research Unit Analytical BioGeoChemistry at Helmholtz Zentrum München

The non-targeted metabolomics activities in the Analytical BioGeoChemistry Research Unit at Helmholtz Zentrum München focusses on profiling metabolites in various environmental and health related host-microbe interactions. The use of innovative non-targeted metabolomics, based on multi-parallel (ultra-) high resolution analytical instruments (UHPLC-qTOF-MS/MS, Bruker solariX 12 Tesla FT-ICR-MS or Bruker cryo-800 MHz NMR spectrometer) allows to get a full insight into the metabolome profile.
High-field magnetic resonance mass spectrometry (Bruker solariX 12 Tesla FT-ICR-MS) screens up to 10.000 mass features with highest mass resolving power in a single analysis with an average of 2.500 annotated metabolites in metabolite databases or massdifference networks.
Furthermore, identification of annotated metabolites can be followed using liquid chromatography based mass spectrometry techniques (Waters Acquity UHPLC or Tandem LC from Thermo Scientific Dionex Ultimate 3000 system coupled to maXis plus UHR-qTOF-MS,) in complementary or in tandem column setups. Complete structural identification of most interesting candidates as well as non-targeted metabotyping is established for NMR spectroscopy.
Raw data processing of MS and NMR is built up since many years and is directly applicable for the projects of miTarget consortium.
Aims
Non-Targeted Metabolomics: We here aim
- To apply LC-MS/MS and NMR based techniques to profile comprehensively stool and serum metabolites in IBD pathology in mice and humans within miTarget
- To provide experimental metabolite data for the validation of metabolic modeling
- To generate and analyze metabolite data for miTarget project partners
- Method development for increased metabolite identification in biological matrices via tandem LC-MS/MS and absolute quantification of stool metabolites via NMR spectroscopy
In close collaboration with various sub-projects of the research unit we will investigate changes in metabolite concentrations in disease as well as in response to perturbations and identify potential biomarkers of therapeutic/prognostic value for IBD pathology.
LC-MS/MS based workflow of non-targeted metabolomics for miTarget Project partners
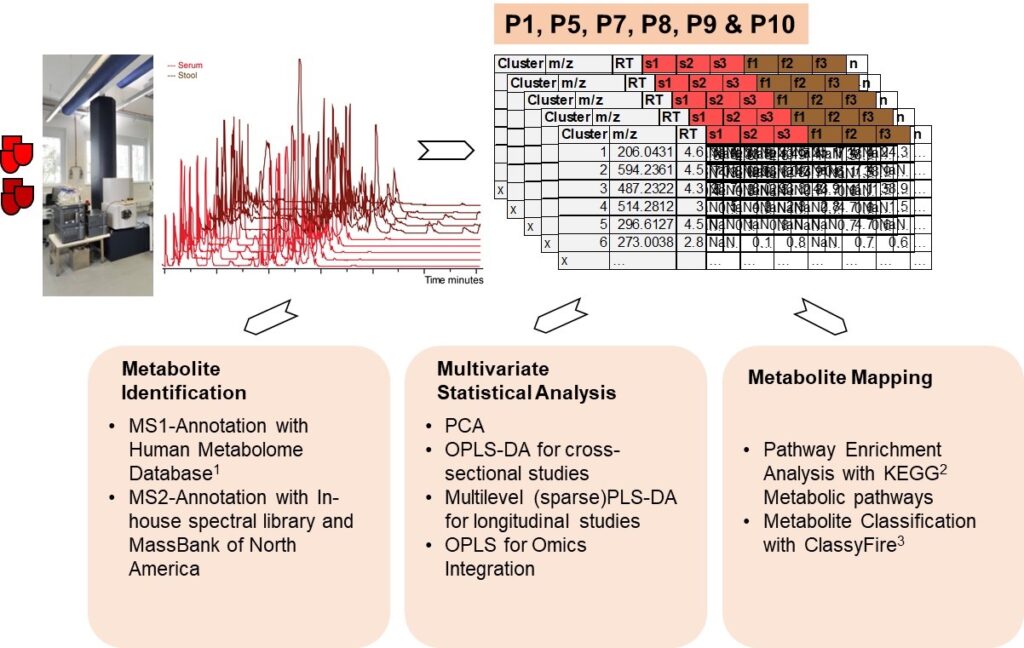
Stool and serum samples from miTarget project partners will be chromatographically separated on a HILIC phase, suitable for polar metabolites. After processing of raw LC-MS/MS data, data with clusters containing mass signals (m/z), retention time (RT), sample names (s1-sn, f1-fn), normalized peak areas are subjected to statistics such as principal component analysis, OPLS-DA for cross-sectional, multilevel (sparse)PLS-DA for lontitudinal studies or OPLS for Omics Integration. Metabolite identification is done on m/z alues by the MS1 annotation with The Human Metabolome Database [1]. The fragmentation of m/z values results in unique MS/MS (MS2) which will be identified by matching against MassBank of North America or in an in-house spectral library, acquired during last years in Analytical BioGeoChemistry at Helmholtz Zentrum München. Identification of metabolites allows to perform further analysis such KEGG pathways analysis [2] or classification of metabolites with ClassyFire [3].
[1] Wishart DS, Tzur D, Knox C, et al., HMDB: the Human Metabolome Database. Nucleic Acids Res. 2007 Jan;35(Database issue):D521-6.
[2] Kanehisa, M. and S. Goto (2000). “KEGG: kyoto encyclopedia of genes and genomes.” Nucleic acids research 28: 27-30.
[3] Djoumbou Feunang Y, Eisner R, Knox C, Chepelev L, Hastings J, Owen G, Fahy E, Steinbeck C, Subramanian S, Bolton E, Greiner R, and Wishart DS. ClassyFire: Automated Chemical Classification With A Comprehensive, Computable Taxonomy. Journal of Cheminformatics, 2016, 8:61.
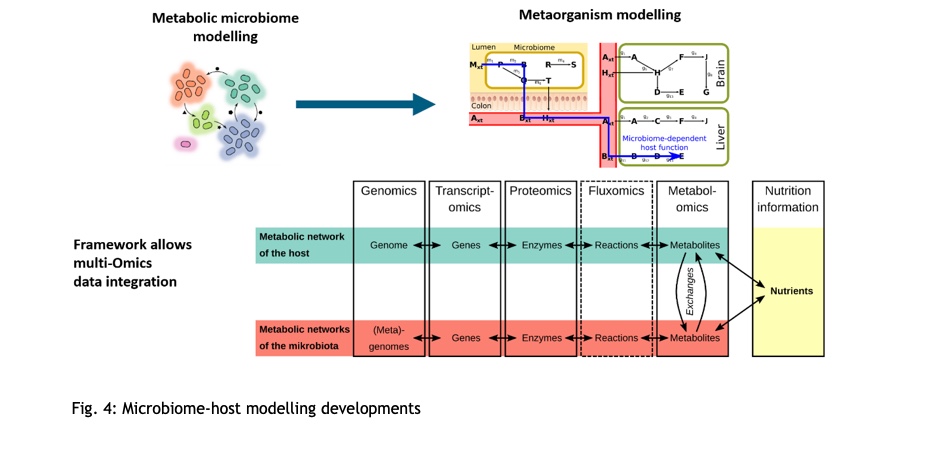
Metabolic modelling: We here aim
- To use and develop microbial community modelling approaches including metabolic metaorganism modelling
- To gain deeper insight into changes in microbial metabolic pathways in the context of IBD using microbial community modelling
- To disentangle microbiome-driven from host-driven components of disease pathology using and refining the concept of metabolic meta-organism models that represent integrated models of microbiome and host
Researchers
Other important members of Z
- Prof. Dr. Dr. Agr. Philippe Schmitt-Kopplin, director of the Institute
- Dr. rer. nat Marianna Lucio, statistician
- Kristina Haslauer, pHD student
- Brigitte Look, technician
- Dr. rer. nat. Silvio Waschina
- Johannes Zimmermann





